
Over the years, many students have had the opportunity to complete their MSc in Computer Science degrees at Brock University. Master's studies offers students a rewarding intellectual experience, with the opportunity to conduct world-class research in a variety of fields in computer science. Research is conducted in close collaboration with faculty supervisors, who pride themselves on being accessible to students for regular meetings. Students also enjoy a warm and friendly social atmosphere with fellow students, faculty and staff. Our student body comprises a diversity of domestic and international students, which greatly contributes to the rich graduate experience available at Brock.
Alumni have found that their MSc degrees have been extremely beneficial for their careers after graduation. An MSc in Computer Science raises the profile of students applying for competitive industry positions. The technical skills and research experiences they acquired have proven valuable for their future careers. After graduation, MSc students have proceeded to careers in industry in Canada, the USA, and overseas. Our graduates are employed a variety of companies, from large, well-known companies (Google, Microsoft,...), to smaller start-ups. Many of our students have remained in the "Golden Horseshoe" area between Toronto and the Niagara peninsula.
Our MSc is also a stepping stone to further graduate studies at the doctoral level. Many students have continued onwards to PhD degrees in Canada and beyond, for example, U of Guelph, Carleton, Queens, U of Toronto, U of Western Ontario, Dalhousie U, U of Central Florida, and others. Careers in industry or academia are options for these students.
The rest of this page highlights some selected research by our graduate students. Interested readers are invited to peruse the faculty web pages for further examples of graduate research activities. We also encourage you to contact faculty for more information about their research and graduate supervision opportunities.
Student: Anthony Awuley
Title of research: Feature Selection Using Age Layered Genetic Programming
Supervisor: Dr Brian Ross
Research fields: computational intelligence, genetic programming
Brief summary: Anthony explored ways to improve the quality of classifiers obtained with genetic programming.
This research presents the FSALPS (Feature Selection Age Layered Population Structure) evolutionary algorithm. FSALPS performs effective feature subset selection and classification of varied supervised learning tasks. FSALPS uses a novel frequency count system to rank features in the GP population based on evolved feature frequencies. The ranked features are translated into probabilities, which are used to control evolutionary processes such as terminal selection for the construction of GP trees. FSALPS continuously refines the feature subset selection process while simultaneously evolving efficient classifiers through a non-converging evolutionary process that favors selection of features with high discrimination of class labels. Comparative experiments on high-dimensional classification problems show that FSALPS dominated canonical GP in evolving smaller but efficient trees with less bloat expressions. Furthermore, FSALPS significantly outperformed canonical GP and ALPS and some reported feature selection strategies in related literature on dimensionality reduction.
Student: Kyle Harrison
Title of research: Network Similarity Measures and Automatic Construction of Graph Models using Genetic Programming
Supervisor: Dr Beatrice Ombuki-Berman
Research fields: computational intelligence, genetic programming
Brief summary: Kyle uses genetic programming to automatically generate complex network models.
A complex network is an abstract representation of an intricate system of interrelated elements where the patterns of connection hold significant meaning. One particular complex network is a social network whereby the vertices represent people and edges denote their daily interactions. Understanding social network dynamics can be vital to the mitigation of disease spread as these networks model the interactions, and thus avenues of spread, between individuals. To better understand complex networks, algorithms which generate graphs exhibiting observed properties of real-world networks, known as graph models, are often constructed. Determining that a graph model of a complex network accurately describes the target network(s) is not a trivial task as the graph models are often stochastic in nature and the notion of similarity is dependent upon the expected behavior of the network. This research examines a number of well-known network properties to determine which measures best allowed networks generated by different graph models, and thus the models themselves, to be distinguished. A proposed meta-analysis procedure was used to demonstrate how these network measures interact when used together as classifiers to determine network, and thus model, (dis)similarity. The analytical results form the basis of the fitness evaluation for a GP system used to automatically construct graph models for complex networks. Genetic programming was used to reproduce existing, well-known graph models as well as a real-world network. Results indicated that the automatically inferred models exemplified functional similarity when compared to their respective target networks. This approach also showed promise when used to infer a model for a mammalian brain network.
Student: Ashkan Entezari Heravi
Title of research: Disease-Gene Association Using Genetic Programming
Supervisor: Dr Sheridan Houghten
Research fields: bioinformatics, computational intelligence
Brief summary: Ashkan used genetic programming techniques to analyze genetic factors associated with known diseases.

Understanding and identifying genetic mutations can play an important role in our health, by making us able to find better diagnosis and therapeutic strategies for these genetic diseases. There is a vast amount of data regarding the human genome available for the analysis and study of genetic defects. This research is an effort to analyze some useful datasets and to apply different techniques to associate genes with genetic diseases. We analyzed the complex network around genetic codes of two diseases: Parkinson's disease and breast cancer. With this information, we generated a ranking for genes, based on their relevance to these diseases. In order to generate these rankings, centrality measures of all nodes in the complex network surrounding the known disease genes of the given genetic disease were calculated. Using genetic programming, all the nodes were assigned scores based on the similarity of their centrality measures to those of the known disease genes. Obtained results showed that this method is successful at finding these patterns in centrality measures and the highly ranked genes are worthy as good candidate disease genes for being studied.
Student: Ethan Jackson
Title of research: L-Fuzzy Relations in Coq
Supervisor: Dr Michael Winter
Research fields: Formal systems, program logics
Brief summary: Ethan uses mathematical modeling for program analysis.
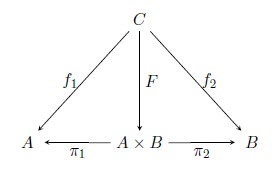
Heyting categories, a variant of Dedekind categories, and Arrow categories provide a convenient framework for expressing and reasoning about fuzzy relations and programs based on those methods. In this research we present an implementation of Heyting and arrow categories suitable for reasoning and program execution using Coq, an interactive theorem prover based on Higher-Order Logic (HOL) with dependent types. This implementation can be used to specify and develop correct software based on L-fuzzy relations such as fuzzy controllers. We give an overview of lattices, L-fuzzy relations, category theory and dependent type theory before describing our implementation. In addition, we provide examples of program executions based on our framework.
Student: Keivan Noorozi
Title of research: On the Hamiltonicity, Connectivity, and Broadcasting Algorithm of the KCube
Supervisor: Dr Ke Qiu
Research fields: algorithms, parallelism
Brief summary: Keivan studied aspects of parallel algorithms. Results of his research can benefit the designs of networks and parallel programs.
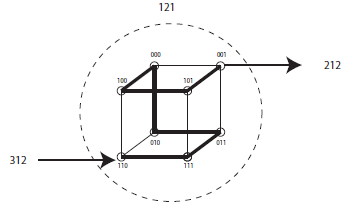
The KCube is a newly proposed topology for connecting many processors in an interconnection or communication network. It combines the well known Kautz graph and the hypercube. The KCube is defined in such a way that it is a class of graphs that have to satisfy two conditions for a graph to be a KCube. Therefore, different versions of the KCube are possible depending on how input and output nodes (vertices) are defined. In this research, a KCube is defined and examined. Further, a KCube graph is proposed that also belongs to the KCube family and is yet different from the original one. Its properties are studied, such as its Hamiltonian1 properties and connectivity. An an optimal broadcasting is also developed.
Student: Xiang Yin
Title of research: Quadtree Representation and Compression of Spatial Data
Supervisor: Dr Ivo Düntsch
Research fields: algorithms, data mining
Brief summary: Xiang proposes a method to improve the compressibility of image data.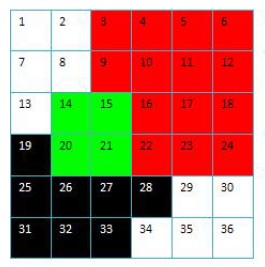
Spatial data representation and compression has become a focus issue in computer graphics and image processing applications. Quadtrees, as one of hierarchical data structures, based on the principle of recursive decomposition of space, always offer a compact and efficient representation of an image. For a given image, the choice of quadtree root node plays an important role in its quadtree representation and final data compression. The goal of this research is to present a heuristic algorithm for finding a root node of a region quadtree, which is able to reduce the number of leaf nodes when compared with the standard quadtree decomposition. The empirical results indicate that, this proposed algorithm has quadtree representation and data compression improvement when in comparison with the traditional method.
